
The disk diffusion test (also known as the agar diffusion test, Kirby–Bauer test, disc-diffusion antibiotic susceptibility test, disc-diffusion antibiotic sensitivity test and KB test) is a culture-based microbiology assay used in diagnostic and drug discovery laboratories. In diagnostic labs, the assay is used to determine the susceptibility of bacteria isolated from a patient's infection to clinically approved antibiotics. This allows physicians to prescribe the most appropriate antibiotic treatment.[1][2][4][5] In drug discovery labs, especially bioprospecting labs, the assay is used to screen biological material (e.g. plant extracts, bacterial fermentation broths) and drug candidates for antibacterial activity. When bioprospecting, the assay can be performed with paired strains of bacteria to achieve dereplication and provisionally identify antibacterial mechanism of action.[6][7]
In diagnostic laboratories, the test is performed by inoculating the surface of an agar plate with bacteria isolated from a patient's infection. Antibiotic-containing paper disks are then applied to the agar and the plate is incubated. If an antibiotic stops the bacteria from growing or kills the bacteria, there will be an area around the disk where the bacteria have not grown enough to be visible. This is called a zone of inhibition. The susceptibility of the bacterial isolate to each antibiotic can then be semi-quantified by comparing the size of these zones of inhibition to databases of information on known antibiotic-susceptible, moderately susceptible and resistant bacteria. In this way, it is possible to identify the most appropriate antibiotic for treating a patient's infection.[1][2] Although the disk diffusion test cannot be used to differentiate bacteriostatic and bactericidal activity, it is less cumbersome than other susceptibility test methods such as broth dilution.[4]
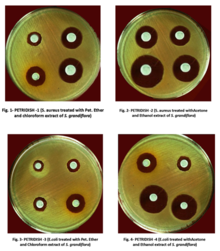
In drug discovery labs, the disk diffusion test is performed slightly differently than in diagnostic labs. In this setting, it is not the bacterial strain that must be characterized, but a test extract (e.g. a plant or microbial extract). The agar plate is therefore inoculated with a bacterial strain of known phenotype (often an ATCC or NCTC strain), and disks containing the test extract are applied to the surface.[6] Zone of inhibition sizes cannot be used as a semi-quantitative measure of antibacterial potency because different extracts contain molecules with different diffusion characteristics (different molecular sizes, hydrophilicities etc.). Zone of inhibition sizes can be used for the purpose of dereplication though. This is achieved by testing each extract against paired strains of bacteria (e.g. streptomycin-susceptible and -resistant strains to identify streptomycin-containing extracts). Paired strains (e.g. wild type and target overexpressing strains) can also be used to identify antibacterial mechanism of action.[6][7]
- ^ a b c EUCAST (January 2021). "Antimicrobial susceptibility testing: EUCAST disk diffusion method" (PDF). www.eucast.org. EUCAST. Retrieved March 16, 2021.
- ^ a b c Brown DF, Kothari D (October 1975). "Comparison of antibiotic discs from different sources". Journal of Clinical Pathology. 28 (10): 779–83. doi:10.1136/jcp.28.10.779. PMC 475859. PMID 1214010.
- ^ Sahu, BK (2013). Antimicrobial properties of aerial part of Sesbania grandiflora (Linn.) (Semester project). The Pharmaceutical College Barpali, India.
- ^ a b Bauer AW, Perry DM, Kirby WM (August 1959). "Single-disk antibiotic-sensitivity testing of staphylococci: An analysis of technique and results". Archives of Internal Medicine. 104 (2): 208–216. doi:10.1001/archinte.1959.00270080034004. PMID 13669774.
- ^ Bauer AW, Kirby WM, Sherris JC, Turck M (April 1966). "Antibiotic susceptibility testing by a standardized single disk method". American Journal of Clinical Pathology. 45 (4): 493–496. doi:10.1093/ajcp/45.4_ts.493. PMID 5325707.
- ^ a b c Cushnie TP, Cushnie B, Echeverría J, Fowsantear W, Thammawat S, Dodgson JL, Law S, Clow SM (June 2020). "Bioprospecting for antibacterial drugs: a multidisciplinary perspective on natural product source material, bioassay selection and avoidable pitfalls". Pharmaceutical Research. 37 (7): Article 125. doi:10.1007/s11095-020-02849-1. PMID 32529587. S2CID 254932190.
- ^ a b Singh SB, Young K, Miesel L (August 2011). "Screening strategies for discovery of antibacterial natural products". Expert Review of Anti-infective Therapy. 9 (8): 589–613. doi:10.1586/eri.11.81. PMID 21819327. S2CID 986144.
